I'm trying to understand the function lmer. I've found plenty of information about how to use the command, but not much about what it's actually doing (save for some cryptic comments here: http://www.bioconductor.org/help/course-materials/2008/PHSIntro/lme4Intro-handout-6.pdf). I'm playing with the following simple example:
library(data.table)
library(lme4)
options(digits=15)
n<-1000
m<-100
data<-data.table(id=sample(1:m,n,replace=T),key="id")
b<-rnorm(m)
data$y<-rand[data$id]+rnorm(n)*0.1
fitted<-lmer(b~(1|id),data=data,verbose=T)
fitted
I understand that lmer is fitting a model of the form Y_{ij} = beta + B_i + epsilon_{ij}, where epsilon_{ij} and B_i are independent normals with variances sigma^2 and tau^2 respectively. If theta = tau/sigma is fixed, I computed the estimate for beta with the correct mean and minimum variance to be
c = sum_{i,j} alpha_i y_{ij}
where
alpha_i = lambda/(1 + theta^2 n_i)
lambda = 1/[\sum_i n_i/(1+theta^2 n_i)]
n_i = number of observations from group i
I also computed the following unbiased estimate for sigma^2:
s^2 = \sum_{i,j} alpha_i (y_{ij} - c)^2 / (1 + theta^2 - lambda)
These estimates seem to agree with what lmer produces. However, I can't figure out how log likelihood is defined in this context. I calculated the probability density to be
pd(Y_{ij}=y_{ij}) = \prod_{i,j}[f_sigma(y_{ij}-ybar_i)]
* prod_i[f_{sqrt(sigma^2/n_i+tau^2)}(ybar_i-beta) sigma sqrt(2 pi/n_i)]
where
ybar_i = \sum_j y_{ij}/n_i (the mean of observations in group i)
f_sigma(x) = 1/(sqrt{2 pi}sigma) exp(-x^2/(2 sigma)) (normal density with sd sigma)
But log of the above is not what lmer produces. How is log likelihood computed in this case (and for bonus marks, why)?
Edit: Changed notation for consistency, striked out incorrect formula for standard deviation estimate.
The links in the comments contained the answer. Below I've put what the formulae simplify to in this simple example, since the results are somewhat intuitive.
lmer fits a model of the form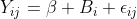 , where
, where 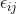 and
and 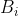 are independent normals with variances
are independent normals with variances 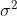 and
and 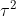 respectively. The joint probability distribution of
respectively. The joint probability distribution of 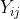 and
and 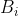 is therefore
is therefore
where
The likelihood is obtained by integrating this with respect to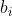 (which isn't observed) to give
(which isn't observed) to give
where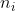 is the number of observations from group
is the number of observations from group 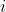 , and
, and 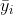 is the mean of observations from group
is the mean of observations from group 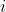 . This is somewhat intuitive since the first term captures spread within each group, which should have variance
. This is somewhat intuitive since the first term captures spread within each group, which should have variance 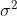 , and the second captures the spread between groups. Note that
, and the second captures the spread between groups. Note that 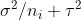 is the variance of
is the variance of 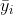 .
.
However, by default (REML=T) lmer maximises not the likelihood but the "REML criterion", obtained by additionally integrating this with respect to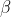 to give
to give
where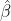 is given below.
is given below.
Maximising likelihood (REML=F)
If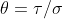 is fixed, we can explicitly find the
is fixed, we can explicitly find the 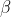 and
and 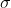 which maximise likelihood. They turn out to be
which maximise likelihood. They turn out to be
Note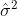 has two terms for variation within and between groups, and
has two terms for variation within and between groups, and 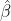 is somewhere between the mean of
is somewhere between the mean of 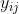 and the mean of
and the mean of 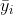 depending on the value of
depending on the value of 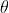 .
.
Substituting these into likelihood, we can express the log likelihood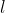 in terms of
in terms of 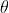 only:
only:
lmer iterates to find the value of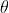 which minimises this. In the output,
which minimises this. In the output, 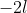 and
and 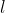 are shown in the fields "deviance" and "logLik" (if REML=F) respectively.
are shown in the fields "deviance" and "logLik" (if REML=F) respectively.
Maximising restricted likelihood (REML=T)
Since the REML criterion doesn't depend on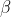 , we use the same estimate for
, we use the same estimate for 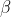 as above. We estimate
as above. We estimate 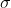 to maximise the REML criterion:
to maximise the REML criterion:
The restricted log likelihood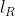 is given by
is given by
In the output of lmer,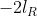 and
and 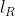 are shown in the fields "REMLdev" and "logLik" (if REML=T) respectively.
are shown in the fields "REMLdev" and "logLik" (if REML=T) respectively.